Are you looking for our Lund University AlphaFold-focussed structure prediction facility LU-Fold? Click here https://atkinson-lab.com/lu-fold/
Atkinson lab research
The broad focus of our research is discovery of new biology through bioinformatic analyses and prediction of protein function and structure. We’re interested in how computational analyses can be combined with experimental research to reconstruct functional evolution of proteins, and to generate testable hypotheses about unknown aspects of protein function.
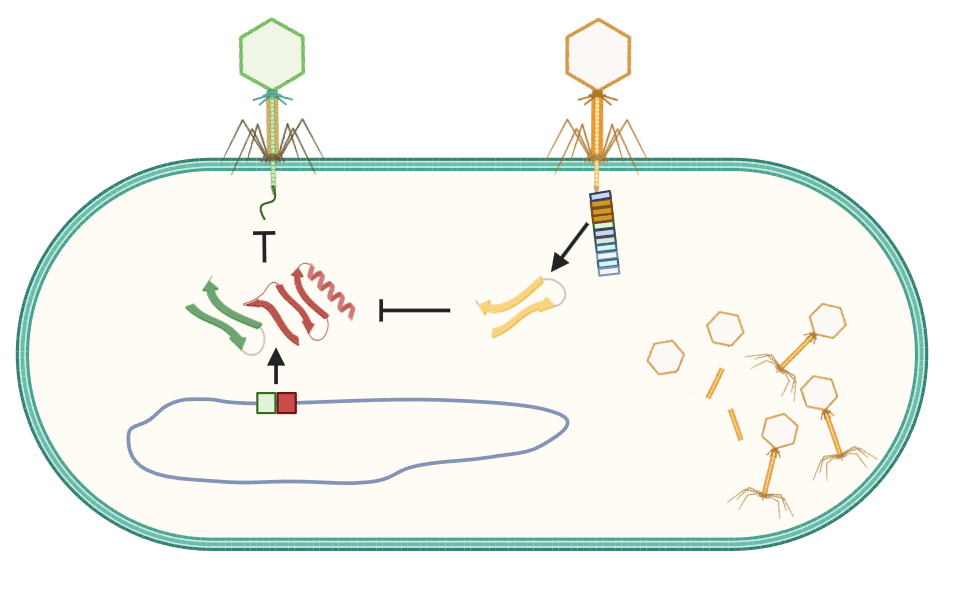
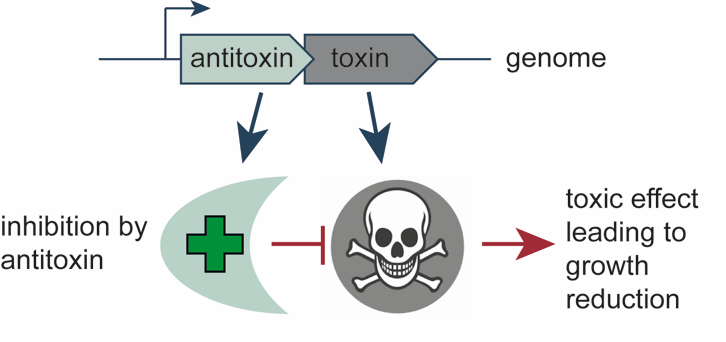
We are particularly interested in proteins involved in microbial attack and defence, such as toxin antitoxin systems and other phage defence mechanisms.
Ongoing projects in the group and relevant publications:
- Prediction and verification of novel toxin-antitoxin systems and other phage defence systems
- Development of bioinformatics tools for comparative evolutionary sequence analysis (see our tools page here https://server.atkinson-lab.com/)
- High throughput structural and functional prediction of proteins and protein complexes
- eg https://pubmed.ncbi.nlm.nih.gov/37556498/ and see our infrastructure here https://atkinson-lab.com/lu-fold/
- Evolution and function of ABCF translation and antibiotic resistance factors in eukaryotes and bacteria
- Functional diversity of small alarmone (ppGpp) synthetase-like enzymes in bacteria and bacteriophages
- Discovery of new ribosome-associating proteins involved in translation, ribosome quality control and antibiotic resistance
- High-throughput AlphaFold predictions of viral proteins and protein-protein interactions
We are part of a joint team with the experimental lab of Vasili Hauryliuk: https://portal.research.lu.se/en/persons/vasili-hauryliuk
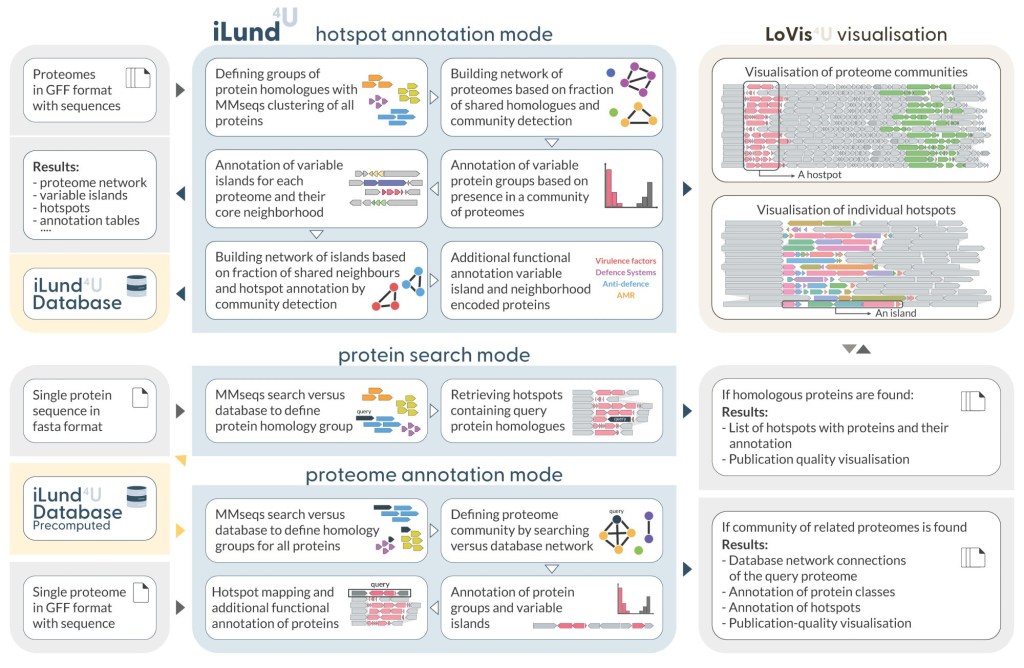
Two of our newest Atkinson lab tools are Lovis4u and iLund4u, made by PhD student Artyom Egorov. Lund4u finds diversity hotspots/islands encoding for example virulence or MGE defence systems www.biorxiv.org/content/10.1…
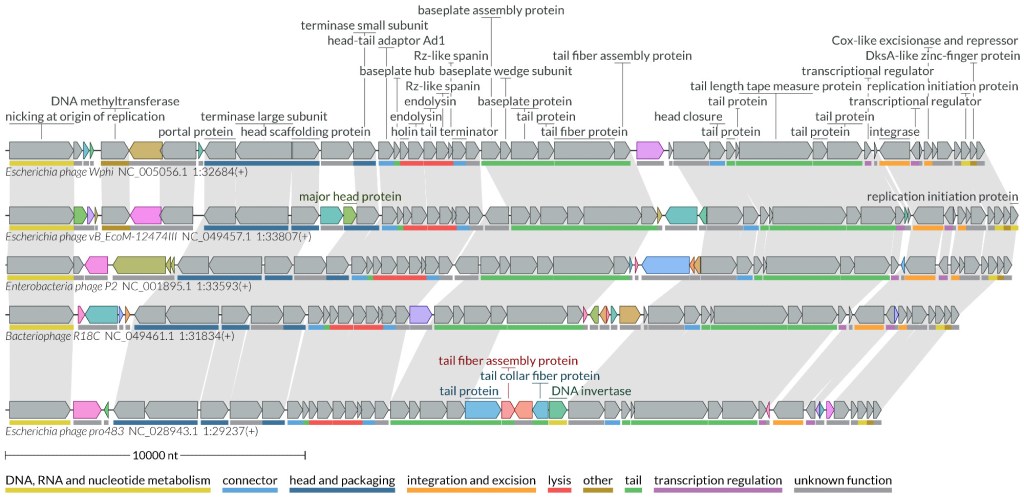
loVis4u sensitively compares & visualises phage or plasmid sequences with beautiful vector graphics www.biorxiv.org/content/10.1… .
For many of our discoveries we have used our tool FlaGs, available via a server here https://server.atkinson-lab.com/webflags
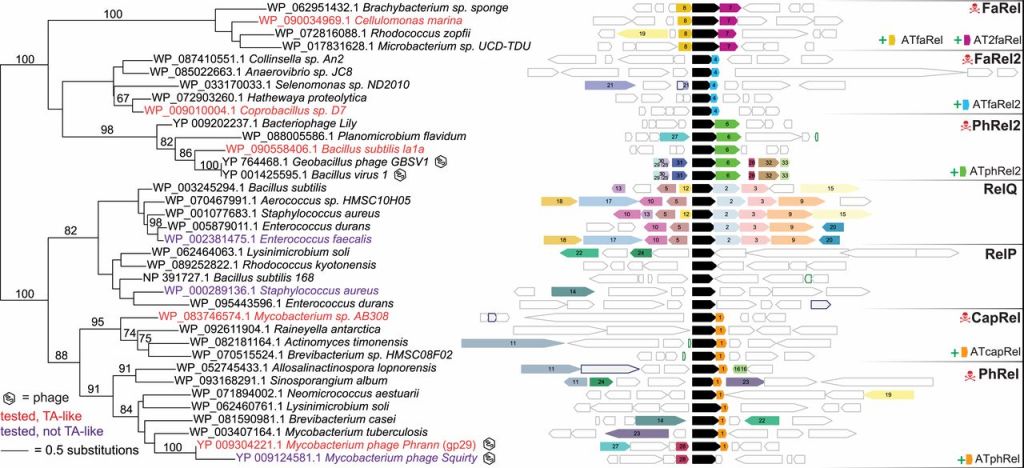
Toxin-antitoxin genes are enigmatic components of microbial genomes. What toxins of TA systems all have in common is that they slam the brakes on growth and reproduction. This is counteracted by their antitoxins which are encoded by adjacent genes. Antitoxins can also work in different ways, either binding to the toxin to stop its action, or counteracting the effect of the toxin in an indirect way. We are exploring the vast zoo of weird and wonderful TA systems in bacterial and bacteriophage genomes, understanding how they work and investigating the biotechnological opportunities they may offer.
I have a long-term interest in GTPase translation factors and how they have evolved in different branches of the tree of life. These core components of translation are at the heart of life itself and were present in the last common ancestor of all life on Earth.