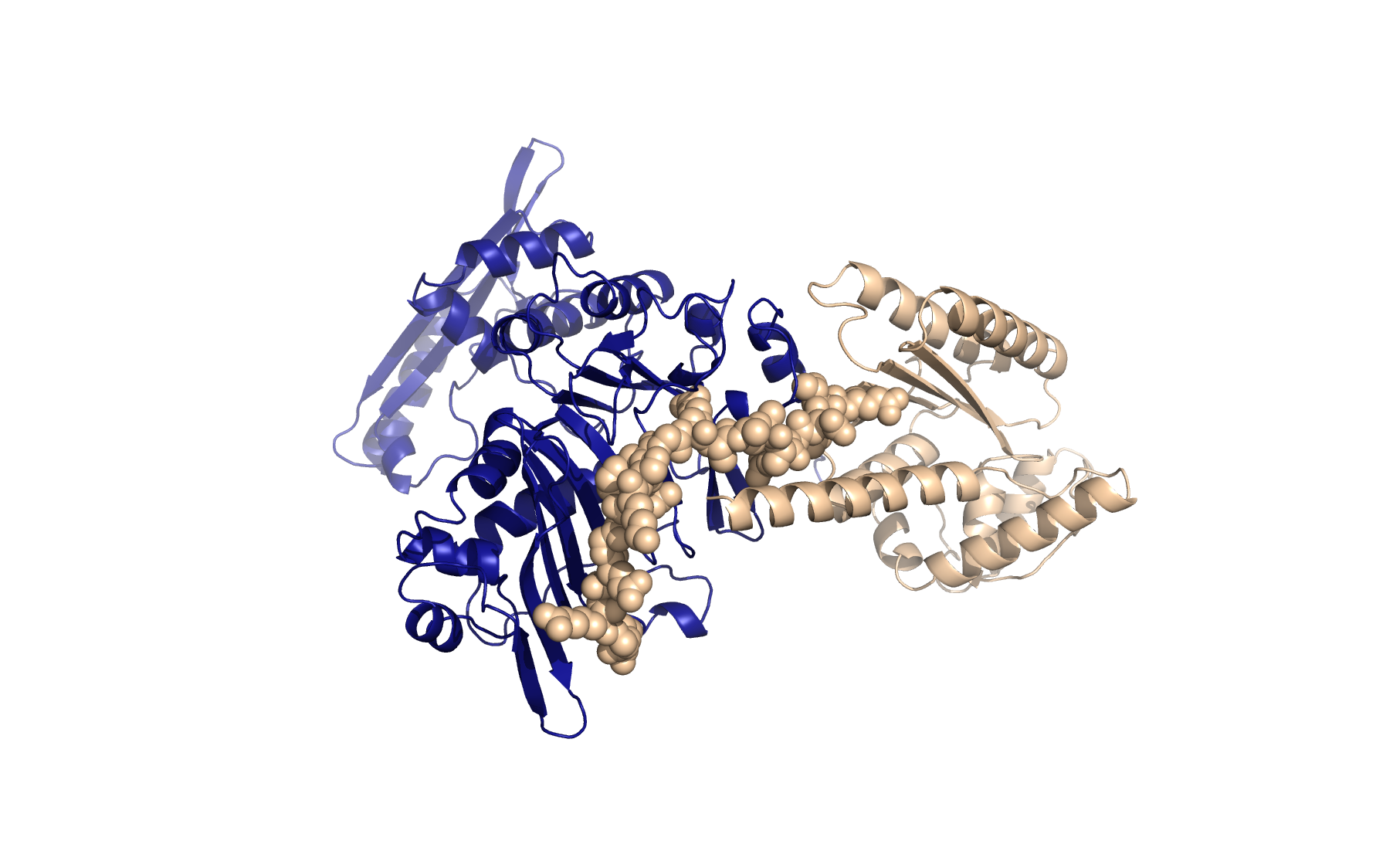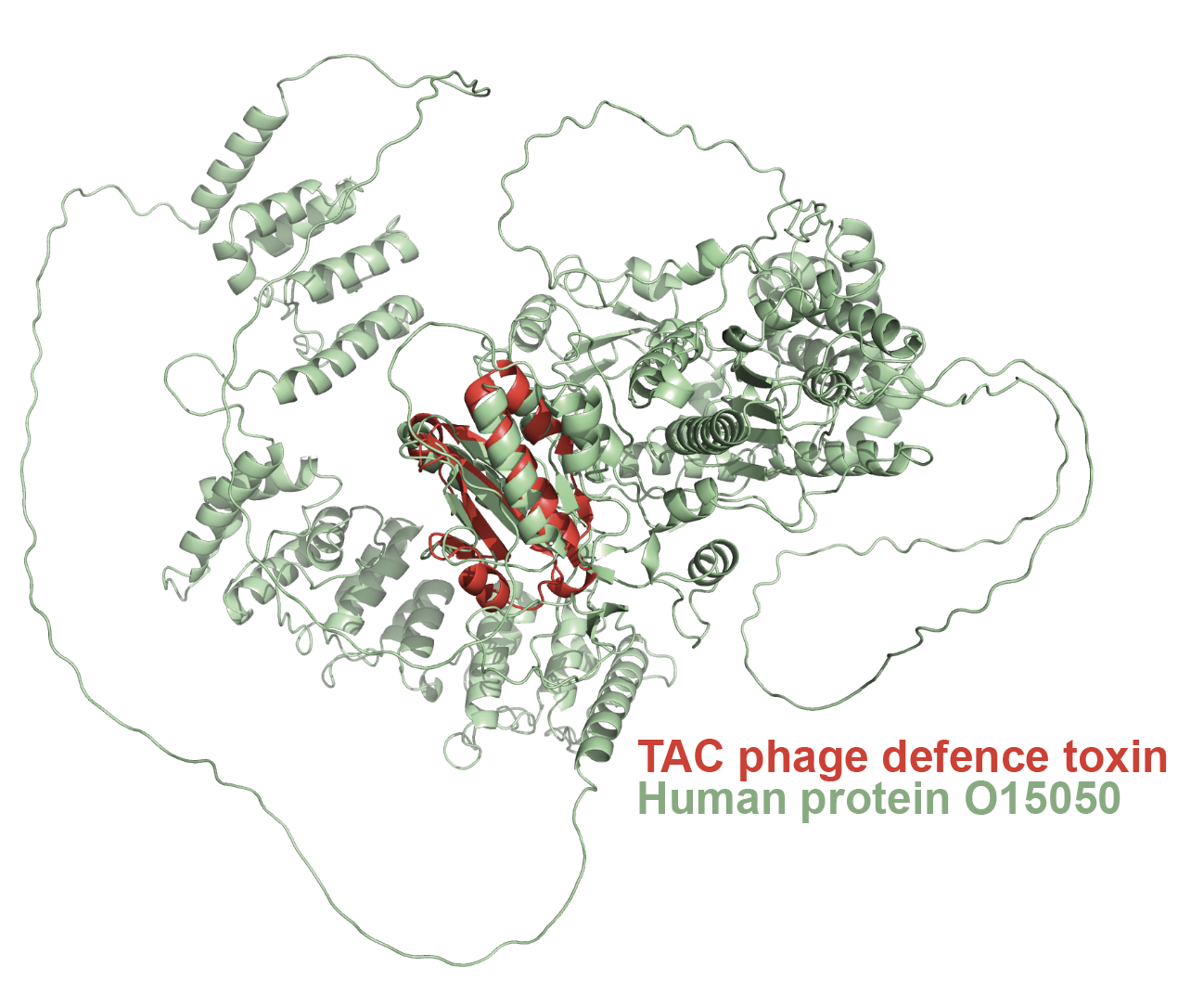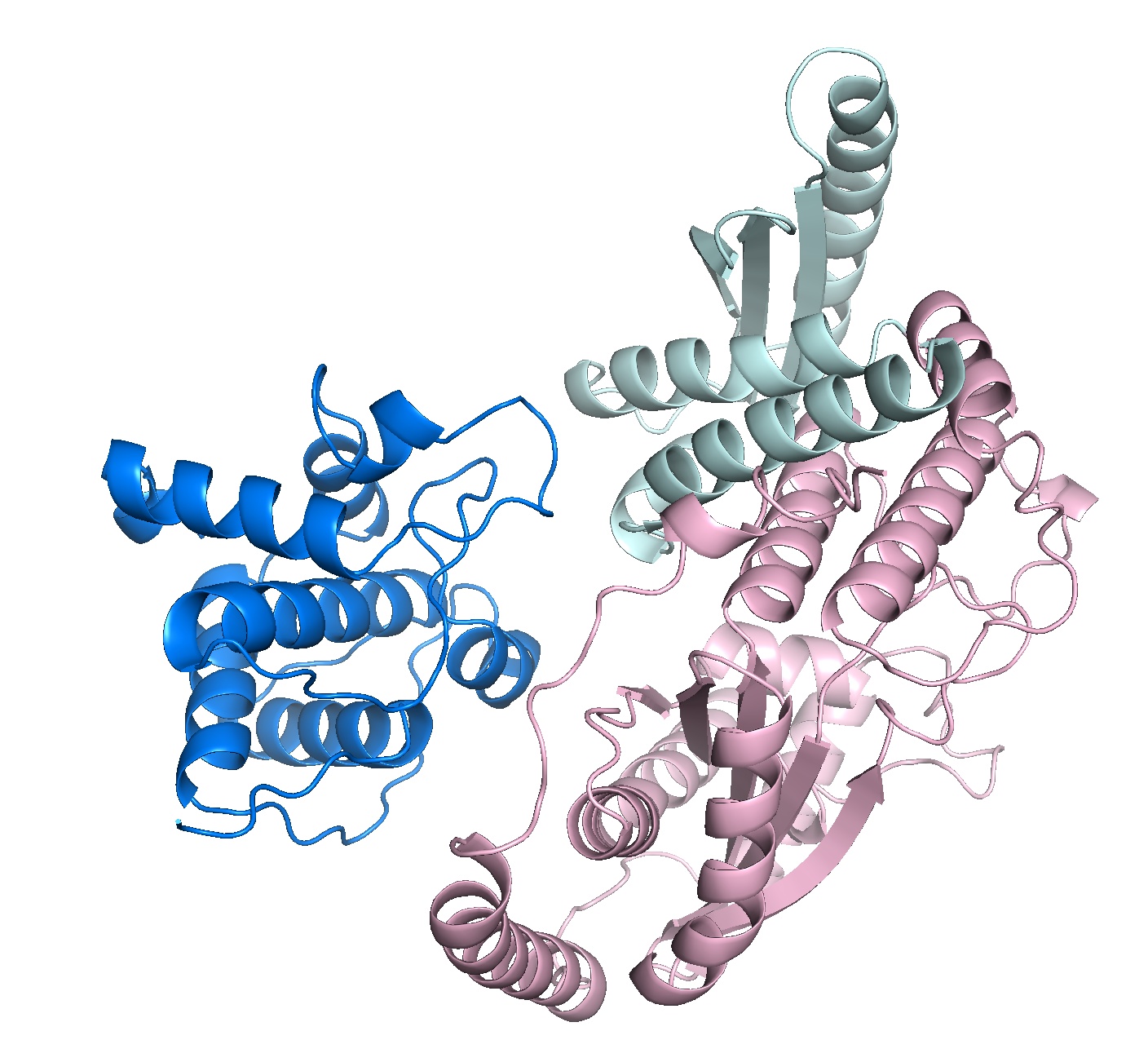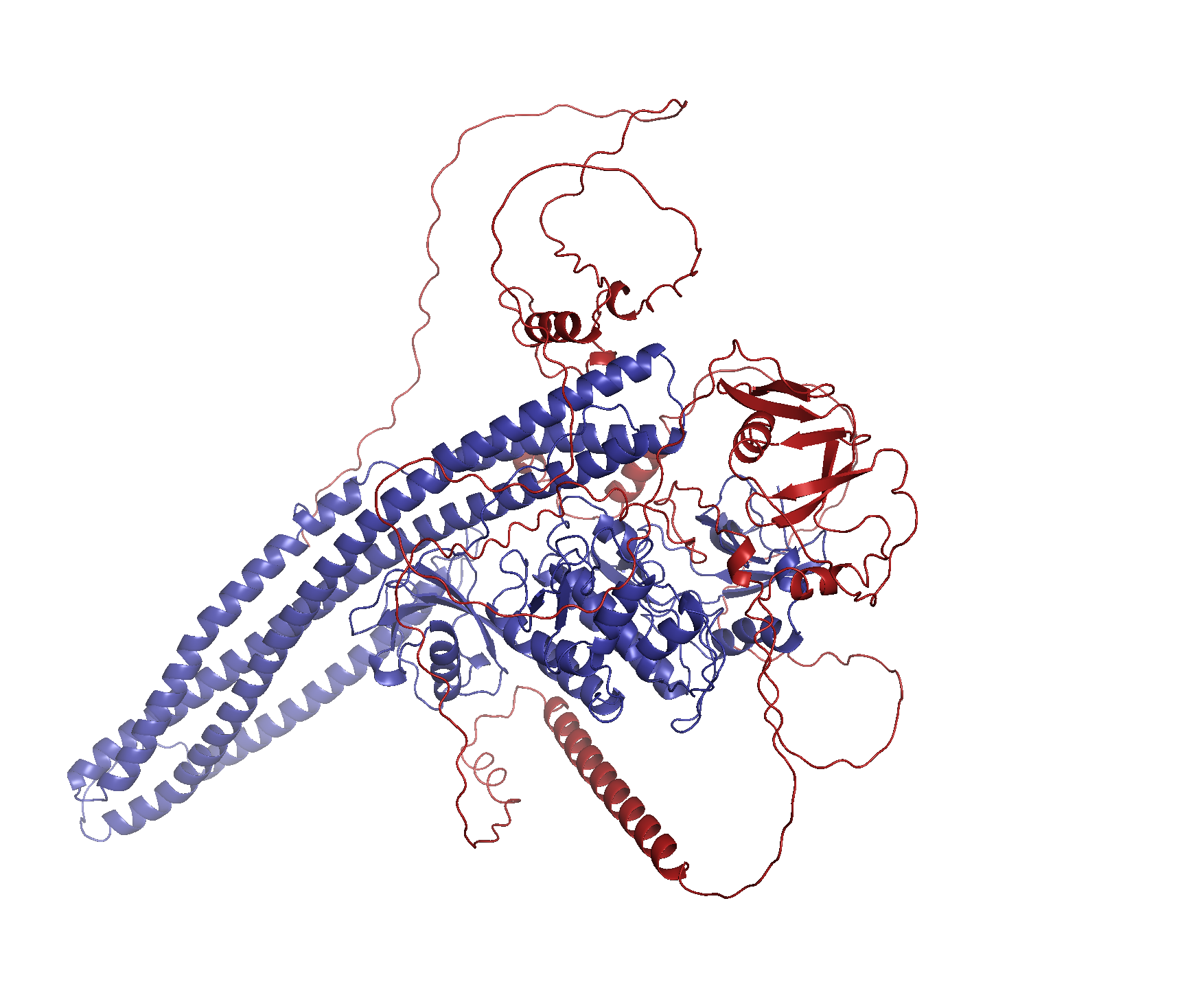
Welcome to LU-Fold!
LU-Fold is a Lund University-based facility for helping researchers predict protein structures of interest using the cutting-edge method AlphaFold2 (Nature Methods method of the year, 2021). LU-Fold specialises in high-throughput prediction of protein complexes to predict novel protein-protein interactions. For example, we can predict pairwise interactions of a protein of interest with all other proteins in a proteome to find new binding partners and molecular binding interfaces. We also offer tutorials, workshops and online guides to help others make their own predictions, compare structures with others in the public domain, visualise results and make publication-quality structures.
We run as a service, accessing national high performance computing infrastructure to make high-throughput structural predictions. Users do not have to have any previous bioinformatics or structural biology experience.
The services we provide include:
- Predicting pairwise binding interactions of a protein of interest with all other proteins in a proteome
- Predicting the structures of all proteins in a proteome (for instance from a newly sequenced genome)
- Predicting higher order structures of larger complexes
- Predicting the effect of mutations and truncations on interactions
- Workshops on using protein structure prediction tools and protein structure visualisation
New for 2025: LU-Fold and the National Bioinformatics Infrastructure Sweden (NBIS) have officially joined to offer improved protein structure prediction services through SciLifeLab.
To apply for support in protein structure prediction, please contact NBIS and LU-Fold here
https://nbis.se/services/bioinformatics/userfee/apply To read more, please visit www.nbis.se and www.medicine.lu.se/research/list-research-infrastructures/lu-fold.
Email gemma.atkinson@med.lu.se to get added to the LU-Fold mailing list.